Quick Guide to Multi-Protein Analysis
Follow this short guide and learn how to get started with Multi-Protein Analysis. including Multi-Protein Quantitation, Multi-Protein Identification, and Multi-Protein Preview workflows.
Multi-Protein Quantitation, Multi-Protein Identification, and Multi-Protein Preview Workflows
The Multi-Protein workflows each have three separate tabs: Samples, Sequences, and Processing nodes. Project Creation steps differ between each workflow. Below outlines the process of Project Creation using the Multi-Protein Identification workflow. Additional details on each workflow can be found in the Byos User Manual.
Create a Workflow
Click to launch any of the above mentioned workflows. The below steps highlight creation using the Multi-Protein Identification workflow.
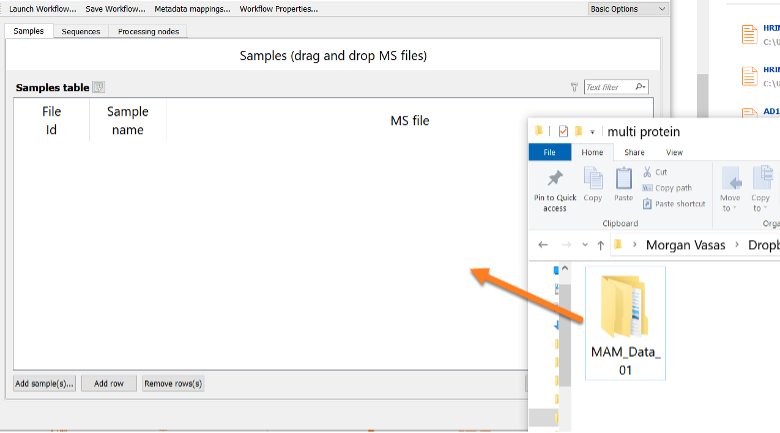
Drag and drop the sample file(s) to be processed into the Samples tab, as shown in the figure below:
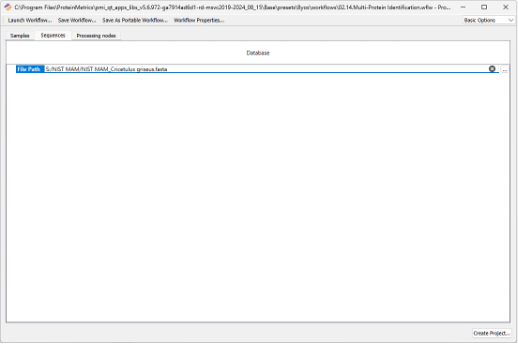
In the Sequences tab, specify the FASTA database:
Either manually enter the enzyme specificity and fixed modification based upon the alkylating reagent used, or simply import the search parameters created by the Multi-Protein Preview analysis
On the Processing tab of the workflow editor, right click on the Multi-Protein Identification node to display the context menu, select Load byparms:
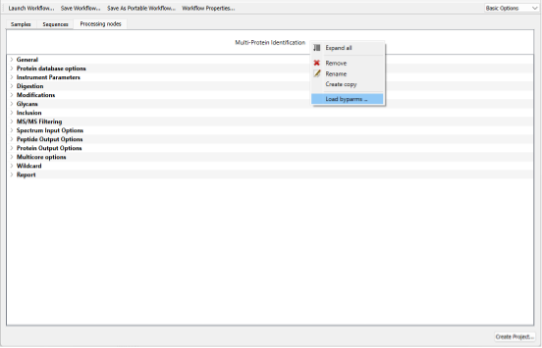
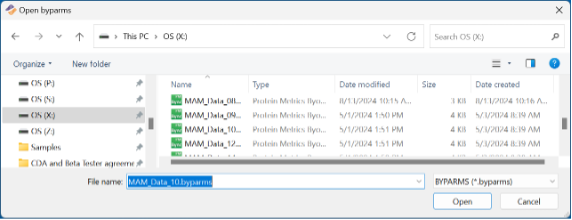
Browse and select the parameter file created by Preview:
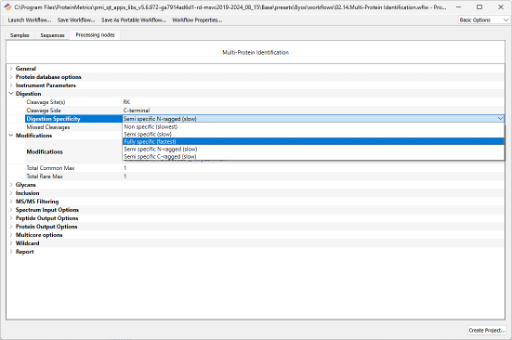
If desired, modify the suggested search parameters:
Browse to a Target output folder and enter a desired project name:
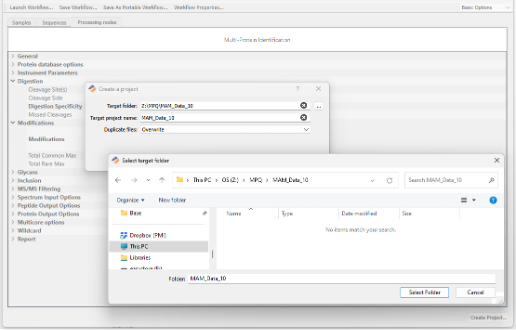
Click Create Project to begin analysis. Depending upon the size of the data and/or fasta databases used, your protein identification results should be available in a relatively short period of time.
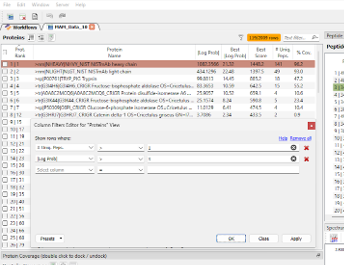
From the open result, apply filters to the Protein view to focus on the highest ranked proteins:
Learn More
Workflow How-Tos
Knowledge Base how to guides for using a variety of workflows.
Byos General
Knowledge Base general articles for using Byos
Application Notes
Application notes on a variety of subjects
Byonic
Knowledge base commonly asked questions about Byonic
More Resources

VIDEO SERIESUsing Byonic
The Byonic tutorials explain MS/MS integration, peptide searches, charge adjustments, filtering PSMs, and report generation.
VIDEO SERIESUsing Byosphere
Byosphere enhances LC-MS analysis with custom searches, dashboards, automated workflows, audit trails, and project management.

WEBINARBovine Milk Immunoglobulin G
Study reveals unique sialylation in bovine colostrum, with potential applications in human infant nutrition.
PUBLICATIONAdvancements in Multi-Protein Quantitation for Host Cell Protein Discovery
Introducing the new multi-protein quantitation mass spectrometric analysis workflow tools for discovery-phase protein scientists.
