De Novo Sequencing of Monoclonal Antibodies
Quickly sequence hundreds or thousands of monoclonal antibodies (mAbs) by mass spectrometry with automatic assembly of individual peptide sequences into the full protein sequence
Explore Workflows
Intact Analysis
Automatic MS/MS spectral annotation, MS level deconvolution, and identification
Oligonucleotides
Characterization and quantitation of oligos
Chromatograms
Align and compare multiple peptide maps to an annotated reference map
Peptide-Level Analysis
Identification of proteins and modified peptides in complex samples
De Novo Sequencing Resources
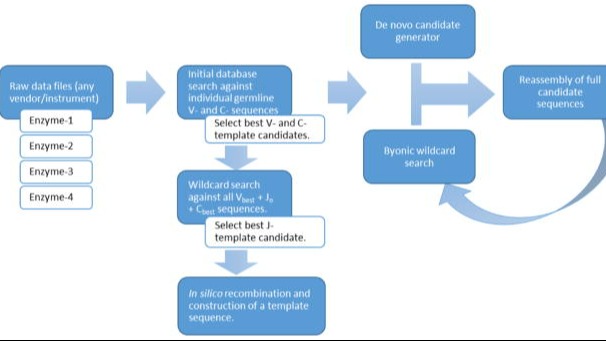
PUBLICATIONAutomated Antibody De Novo Sequencing and Its Utility in Biopharma Discovery
A novel method to automatically de novo sequence antibodies using mass spec and Byos, and a demonstration of algorithm robustness.
WEBINARSARS-CoV-2 Spike Ectodomain
Dive into HDX MS analysis of SARS-CoV-2 spike protein, focusing on glycopeptide inclusion and its challenges.

WEBINAREnhancing Polyclonal IgG Product Characterization
CSL explore uses of Byosphere in characterizing polyclonal IgG products focusing on plasma product development.

WEBINARDashboards By Deep Query
Use dashboards and deep query tools to analyze data, spot outliers, and enhance lab collaboration globally.




