Our Customers, Patents and Publications
See why the world’s leading research organizations are choosing Protein Metrics for their biotherapeutic analysis, reducing the need for time-consuming manual analysis, and gaining deeper insights and more accurate results.
Customers
Trusted by over 500 of the world’s leading biopharma and academic research organizations


















We really feel that better data consistency leads to better business decisions. Byosphere has really been key for us in that regard.
Patents
Protein Metrics has built a powerful suite of products from groundbreaking patents
US 12,352,757 Pseudo-electropherogram construction from peptide level mass spectrometry data
US 12,224,169 Interactive analysis of mass spectrometry data
US 12,205,331 Data compression for multidimensional time series
US 12,040,170 methods and apparatuses for deconvolution of mass spectrometry data
US 12,038,444 Pseudo-electropherogram construction from peptide level mass spectrometry data
US 11,908,130 Apparatuses and methods for digital pathology
US 11,790,559 Data compression for multidimensional time series
US 11,728,150 Methods and apparatuses for determining intact mass of large molecules from mass spectrographic data
US 11,640,901 Methods and apparatuses for deconvolution of mass spectrometry data
US 11,626,274 Interactive analysis of mass spectrometry data including peak selection and dynamic labeling
US 11,346,844 Intact mass reconstruction from peptide level data and facilitated comparison with experimental intact observation
US 11,289,317 Interactive analysis of mass spectrometry data
US 11,276,204 Data compression for multidimensional time series data
US 11,127,575 Methods and apparatuses for determining the intact mass of large molecules from mass spectrographic data
US 10,991,558 Interactive analysis of mass spectrometry data including peak selection and dynamic labeling
US 10,879,057 Interactive analysis of mass spectrometry data
US 10,665,439 Methods and apparatuses for determining the intact mass of large molecules from mass spectrographic data
US 10,546,736 Interactive analysis of mass spectrometry data including peak selection and dynamic labeling
US 10,510,521 Interactive analysis of mass spectrometry data
US 10,354,421 Apparatus and methods for annotated peptide mapping
US 10,319,573 Methods and apparatuses for determining the intact mass of large molecules from mass spectrographic data
US 10,199,206 Interactive analysis of mass spectrometry data
US 9,859,917 Enhanced Data Compression For Sparse Multidimensional Ordered Series Data
US 9,671,408 Wild-card-modification search technique for peptide identification
US 9,640,376 Interactive analysis of mass spectrometry data
US 9,571,122 Enhanced data compression for sparse multidimensional ordered series data
US 9,385,751 Enhanced data compression for sparse multidimensional ordered series data
Published Articles
Protein Metrics has published multiple peer-reviewed articles discussing the unique features, algorithms and workflows used in Byonic, Byos and Byosphere

PUBLICATIONParsimonious Charge Deconvolution for Native Mass Spectrometry
A new “parsimonious” charge deconvolution algorithm well-suited to high-resolution native MS of intact glycoproteins and protein complexes.

PUBLICATIONOrthogonal Comparison of Analytical Methods by Theoretical Reconstruction from Bottom-up Assay Data
Bottom-up methods like peptide mapping can be used to build reconstructions of both theoretical intact mass spectra and theoretical electropherograms.

PUBLICATIONTwo-Dimensional Target Decoy Strategy for Shotgun Proteomics
New target-decoy strategy estimates and controls PSM and protein FDRs simultaneously, regardless of the relative numbers of spectra and proteins.

PUBLICATIONPeak Filtering, Peak Annotation, and Wildcard Search for Glycoproteomics
Improvements to Byonic, a glycoproteomics search program, that addresses the analytical and bioinformatics challenges glycopeptide analysis poses.

PUBLICATIONPreview: A Program for Surveying Shotgun Proteomics Tandem Mass Spectrometry Data
Preview analyzes a set of mass spectra for mass errors, digestion specificity, and known and unknown modifications to facilitate parameter selection.

PUBLICATIONByonic: Advanced Peptide and Protein Identification Software
Byonic is a new software package for peptide and protein identification by tandem mass spectrometry, facilitating a wide range of search possibilities
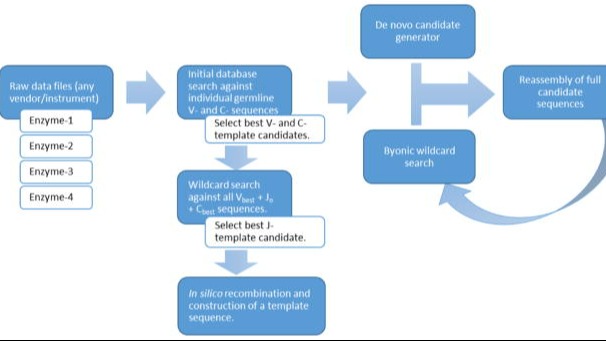
PUBLICATIONAutomated Antibody De Novo Sequencing and Its Utility in Biopharma Discovery
A novel method to automatically de novo sequence antibodies using mass spec and Byos, and a demonstration of algorithm robustness.
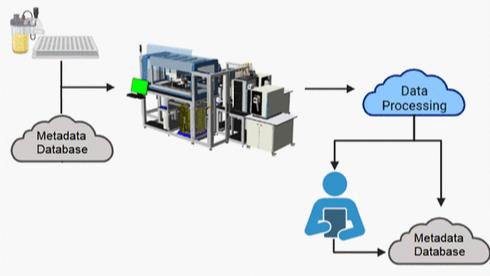
PUBLICATIONLab of the Future: Fully Automated System for High-Throughput Mass Spec Analysis
A state-of-the-art, integrated, multi-instrument automated system for MS characterization of biotherapeutics.
Cited Publications
Read a selection of the hundreds of papers citing Protein Metrics published each year
Poskute, R, et al. "Identification and quantification of chain-pairing variants or mispaired species of asymmetric monovalent bispecific IgG1 monoclonal antibody format using reverse-phase polyphenyl chromatography coupled electrospray ionization mass spectrometry" J Chromatogr B Analyt Technol Biomed Life Sci. 2024. doi: 10.1016/j.jchromb.2024.124085.
Peng, W, et al. "Mass Spectrometry-Based De Novo Sequencing of Monoclonal Antibodies Using Multiple Proteases and a Dual Fragmentation Scheme" J Proteome Res. 2021 Jul 2;20(7):3559-3566. doi: 10.1021/acs.jproteome.1c00169.
Parker, R, et al. "Mapping the SARS-CoV-2 spike glycoprotein-derived peptidome presented by HLA class II on dendritic cells" Cell Rep. 2021 May 25;35(8):109179. doi: 10.1016/j.celrep.2021.109179.
Inoue, K, et al. "Determination of drug-to-antibody ratio of antibody-drug conjugate in biological samples using microflow-liquid chromatography/high-resolution mass spectrometry" Bioanalysis. 2022 Dec;14(24):1533-1545. doi: 10.4155/bio-2022-0219.
Cadang, L, et al. "A Highly Efficient Workflow for Detecting and Identifying Sequence Variants in Therapeutic Proteins with a High Resolution LC-MS/MS Method" Molecules. 2023 Apr 12;28(8):3392. doi: 10.3390/molecules28083392.
Grunert, I, et al. "Detailed Analytical Characterization of a Bispecific IgG1 CrossMab Antibody of the Knob-into-Hole Format Applying Various Stress Conditions Revealed Pronounced Stability" ACS Omega. 2022 Jan 19;7(4):3671-3679. doi: 10.1021/acsomega.1c06305.
Huang, R.Y-C, et al. "Higher-Order Structure Characterization of NKG2A/CD94 Protein Complex and Anti-NKG2A Antibody Binding Epitopes by Mass Spectrometry-Based Protein Footprinting Strategies" J Am Soc Mass Spectrom. 2021 Jul 7;32(7):1567-1574. doi: 10.1021/jasms.0c00399.
Niu, B, et al. "Using New Peak Detection to Solve Sequence Variants Analysis Challenges in Bioprocess Development" J Am Soc Mass Spectrom. 2023 Mar 1;34(3):401-408. doi: 10.1021/jasms.2c00292.
Kristensen, DB, et al. "Optimized Multi-Attribute Method Workflow Addressing Missed Cleavages and Chromatographic Tailing/Carry-Over of Hydrophobic Peptides" Anal Chem. 2022 Dec 13;94(49):17195-17204. doi: 10.1021/acs.analchem.2c03820.
Wu, Z, et al. "High-sensitivity and high-resolution therapeutic antibody charge variant and impurity characterization by microfluidic native capillary electrophoresis-mass spectrometry" J Pharm Biomed Anal. 2023 Jan 20:223:115147. doi: 10.1016/j.jpba.2022.115147.
