Quick Guide to Peptide Analysis
Follow this short guide and learn how to get started with Peptide analysis, including HCP, HotSpot, PTM, PTM (in-silico), SVA (C57)1, SVA (C58)1, S-S, MAM New Peak Detection, System Suitability, and Oxidative Footprinting workflows.
HCP, HotSpot, PTM, SVA (C57) – Specific, SVA (C58) – Specific, S-S, MAM New Peak Detection, System Suitability, Oxidative Footprinting, and PTM (in-silico)
Peptide Analysis workflows have three separate tabs: Samples, Sequences, and Processing nodes.
NOTE: Considerations specific to the HCP and System Suitability workflows in the Sequences tab are called out.
Create a Workflow
Click to launch any of the mentioned workflows. In the figure below, the Post-Translational Modifications (PTM) workflow is highlighted.
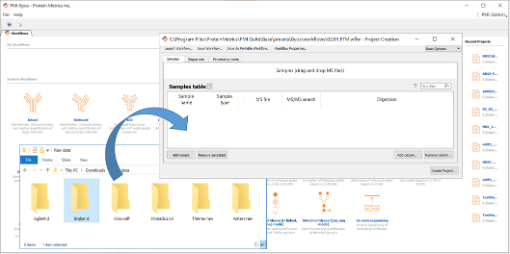
Drag and drop the sample data file(s) to be processed into the Samples tab, as shown in the figure below. These can be from any vendor.
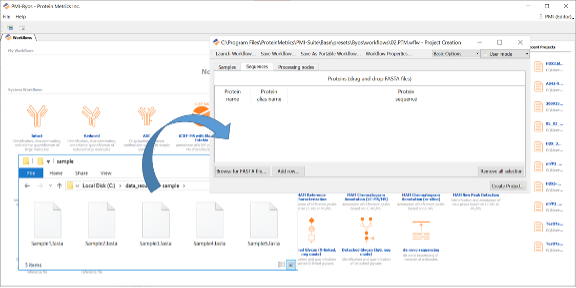
Drag and drop the FASTA file to be processed into the Sequences tab.
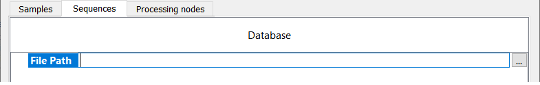
NOTE: In the HCP workflow, the user must instead point to a protein database. The user can either drag and drop as shown in the figure below or direct Byos to a database file. This is done by clicking in the space to activate the light blue “…” box. The user is then prompted to select a file as shown in the figure below.
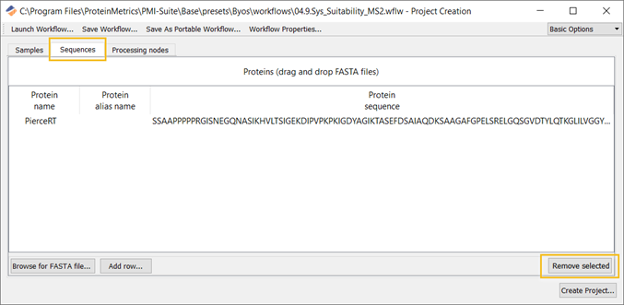
NOTE: In the System Suitability workflow, the Sequences tab is populated by default to include the Pierce RT standard. The user can replace this with another standard if desired. The user can change this by clicking to highlight the line and clicking Remove selection (shown in the figure below). Another protein can be brought in using drag and drop (shown in Figure 3).
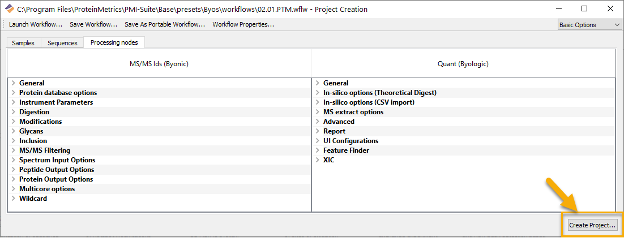
Click Create Project to start processing and to generate the report.
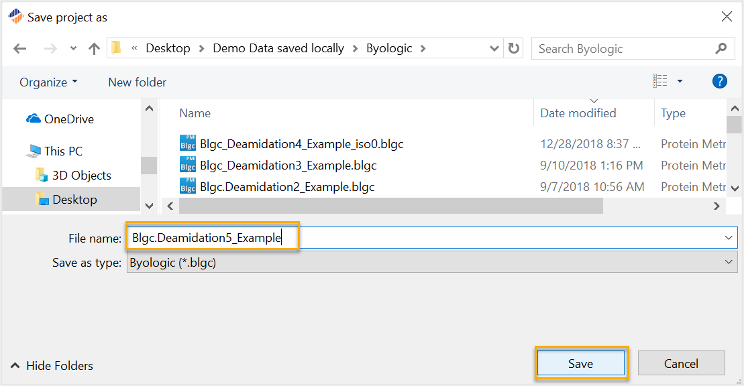
The user will be prompted to name and save the project file. Please note Byos will save this type of project as a *.blgc file.
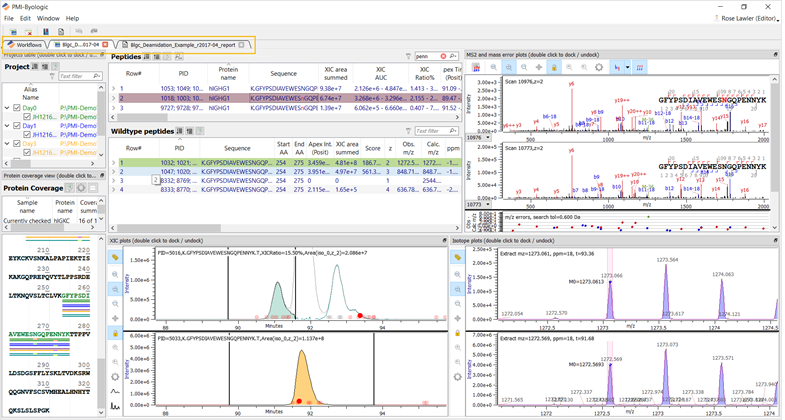
Processing will begin. Once complete, the project and associated report will appear as additional tabs after the Workflows tab, as shown in the figure below.
Learn More
Peptide How-Tos
Knowledge Base commonly asked questions about peptide workflows
Byonic: Focused Protein Databases
Application note focused on protein databases
Byonic: N-Linked Glycopeptide Analysis
Application note on N-linked glycopeptide analysis
Peptide Insights
A selection of Insight articles that cover topics on peptides
More Resources

VIDEO SERIESWorking with Peptide Workflows
Explore peptide workflows, set up searches, interpret scores, validate peptides, manage disulfide mapping, and create reports.

VIDEO SERIESResearch Spotlight on Peptides
Watch these spotlight videos on peptides covering methods for mapping, quantitation, glycosylation, and antibody stability.

WEBINARReconstructing cIEF Profiles Using Peptide Mapping Data
This webinar explores cutting-edge methods for reconstructing cIEF profiles using peptide mapping data with a focus on practical applications.

WEBINARImprovements in Automated Quantitation
Enhance in silico quantification with automated peak detection and signal processing to improve accuracy.
