Monitoring Host-Cell Proteins: A Comparative Analysis
A comparative analysis of MS/MS search algorithms in label-free shotgun proteomics for monitoring host-cell proteins
Webinar Summary
Monitoring host cell proteins (HCPs) is crucial for ensuring product safety and quality in biopharmaceutical development. In this webinar, Somar presents the application of label-free shotgun proteomics for HCP monitoring, benchmarking six prominent MS/MS search tools—Mascot, MaxQuant, SpectroMine, FragPipe, Byos, and PEAKS—utilizing trapped ion mobility spectrometry and ddaPASEF. The study employs Bayesian inference as a robust framework for evaluating tool performance, incorporating Markov Chain Monte Carlo modeling, Bayesian FDR control, and decision theory to balance differential abundance accuracy with uncertainty.
The webinar will highlight the strengths and trade-offs of each search tool, focusing on key metrics such as peptide and protein identifications, data extraction precision, fold change accuracy, linearity, and quantification trueness.
More Resources

VIDEO SERIESResearch Spotlight on Peptides
Watch these spotlight videos on peptides covering methods for mapping, quantitation, glycosylation, and antibody stability.

VIDEO SERIESWorking with Peptide Workflows
Explore peptide workflows, set up searches, interpret scores, validate peptides, manage disulfide mapping, and create reports.

PUBLICATIONByonic: Advanced Peptide and Protein Identification Software
Byonic is a new software package for peptide and protein identification by tandem mass spectrometry, facilitating a wide range of search possibilities
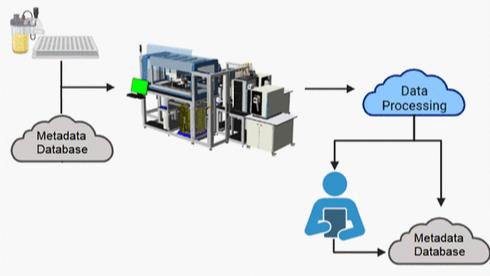
PUBLICATIONLab of the Future: Fully Automated System for High-Throughput Mass Spec Analysis
A state-of-the-art, integrated, multi-instrument automated system for MS characterization of biotherapeutics.














