Resources
Explore our educational resources to advance your knowledge of Protein Metrics
Intact Mass

GUIDEQuick Start Guide to Intact Analysis
An easy guide to getting started with intact, reduced, ADC, intact reconstruction, and IntaBio icIEF-MS workflows.

VIDEO SERIESWorking with Intact Mass Workflows
Run intact mass workflows in Byos. Manage data, sequences, masses, deconvolution, and generate reports for ADCs, glycosylation, and oligos.

VIDEO SERIESResearch Spotlight on Intact Mass
Watch this video on a novel serum stability assay, enhancing accuracy in biologics screening with in vitro methods.

WEBINARIntact Characterization of Multispecifics
Learn about multispecific antibodies and the use of 2D LC-MS for improved production and analysis.

WEBINARAutomating QC in Discovery Therapeutics
Discover how automating high-throughput LC-MS revolutionizes biotherapeutic discovery with faster QC and molecule diversity.

PUBLICATIONOrthogonal Comparison of Analytical Methods by Theoretical Reconstruction from Bottom-up Assay Data
Bottom-up methods like peptide mapping can be used to build reconstructions of both theoretical intact mass spectra and theoretical electropherograms.
Oligonucleotides

GUIDEQuick Guide to Oligonucleotide Analysis
Learn how to get started with oligonucleotide analysis, including digested oligonucleotide analysis.

VIDEO SERIESWorking with Oligonucleotide Workflows
Learn how to automate oligonucleotide data processing, handle raw files, define sequences, and generate custom reports.

VIDEO SERIESWorking with Digested Oligo Workflows
Learn Byos Digested Oligonucleotide workflows, modification setup, investigation tools, and custom report creation.

WEBINARMS Analysis of Intact Oligonucleotides
Explore advanced workflows for oligonucleotide analysis, focusing on fragmentation and MS2 annotation.
Multi-Protein Quantitation

GUIDEQuick Guide to Multi-Protein Analysis
An easy guide to getting started with multi-protein quantitation, multi-protein identification, and multi-protein preview workflows.

VIDEO SERIESWorking with Multi-Protein Quantitation
Rapidly analyze MS data to quantify proteins, manage workflows, and validate peptides in complex samples.

EVENTCASSS MASS SPEC 2024
Read posters about our new Multi-Protein Quantitation workflow and other cutting-edge mass spec advancements and biopharmaceutical innovations.
Peptide

GUIDEQuick Guide to Peptide Analysis
Getting started with HCP, HotSpot, PTM, SVA, S-S, MAM new peak detection, system suitability, and oxidative footprinting workflows.

VIDEO SERIESWorking with Peptide Workflows
Explore peptide workflows, set up searches, interpret scores, validate peptides, manage disulfide mapping, and create reports.

VIDEO SERIESWorking with Disulfides
Learn how to manage disulfide workflows, add samples, define sequences, create disulfide bridge and validate linked peptides.

VIDEO SERIESResearch Spotlight on Peptides
Watch these spotlight videos on peptides covering methods for mapping, quantitation, glycosylation, and antibody stability.

VIDEO SERIESResearch Spotlight on Disulfides
Watch these spotlight videos on rapid cross-link detection, bacterial microcompartment assembly, and bond stability in proteins.

VIDEO SERIESResearch Spotlight on Glycopeptides
Watch these spotlight videos on glycosylation analysis techniques, covering protein glycan structures and SARS-CoV-2 applications.
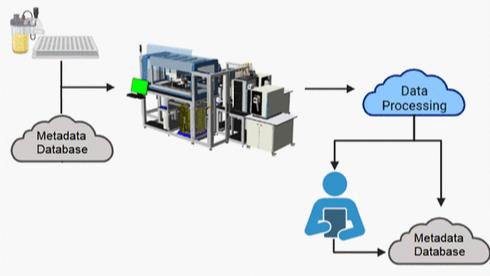
PUBLICATIONLab of the Future: Fully Automated System for High-Throughput Mass Spec Analysis
A state-of-the-art, integrated, multi-instrument automated system for MS characterization of biotherapeutics.
Chromatogram

GUIDEQuick Guide to Chromatogram Analysis
An easy guide to getting started with chromatography workflows, including reference, characterization and in-silico chromatography.

VIDEO SERIESWorking with Chromatogram Workflows
Learn to use chromatogram workflows for peak analysis, adjust baselines, integrate peaks, align traces, and explore glycan analysis.

VIDEO SERIESWorking with Released Glycan Workflows
Learn to use released glycan workflows for N-glycans, O-glycans, and IGG glycans. Explore Glycan cartoons for visual representation.

GUIDEQuick Guide to Released Glycan Analysis
Getting started with released glycan (N-linked), released glycan (O-linked), and released glycan (IgG) workflows.

WEBINARBiosimilars and Innovator Glycans
Compare biosimilar and innovator glycan analysis to understand therapeutic efficacy and patient outcomes.
Reconstruction

GUIDEQuick Guide to Charge Variant Analysis
Follow this short guide and learn how to get started with change variant analysis with reconstruction.

VIDEO SERIESWorking with Reconstruction Workflows
Perform intact reconstruction and charge variant analysis. Set up projects, configure processing nodes, and align CIEF traces effectively.

VIDEO SERIESResearch Spotlight on Multispecifics
The latest in multispecifics with videos on bispecific antibody stability, mispairing, and effective characterization methods.

WEBINARIntact Characterization of Multispecifics
Learn about multispecific antibodies and the use of 2D LC-MS for improved production and analysis.
Higher Order Structure

GUIDEQuick Guide to HDX Workflows
An easy guide to getting started with HDX analysis, including all our HDX workflows.

VIDEO SERIESWorking with Hydrogen Deuterium Exchange (HDX)
The HDX tutorial series explains how to set up HDX workflows, analyze deuterium uptake, and generate detailed protein reports.
VIDEO SERIESResearch Spotlight on Higher Order Structure
Explore higher-order structure in these videos on plasma-induced modification and bacterial microcompartment assembly insights.
WEBINARSARS-CoV-2 Spike Ectodomain
Dive into HDX MS analysis of SARS-CoV-2 spike protein, focusing on glycopeptide inclusion and its challenges.
Other Workflows

GUIDEQuick Guide to HRIM Analysis
Follow this short guide and learn how to get started with MOBILion High-Resolution Ion Mobility (HRIM) analysis.

GUIDEQuick Guide to De Novo Sequence Analysis
An easy guide to getting started with de novo sequence analysis workflows.

VIDEO SERIESResearch Spotlight on De Novo
This video series covers research on advanced mass spectrometry techniques for accurate antibody and protein structure analysis.
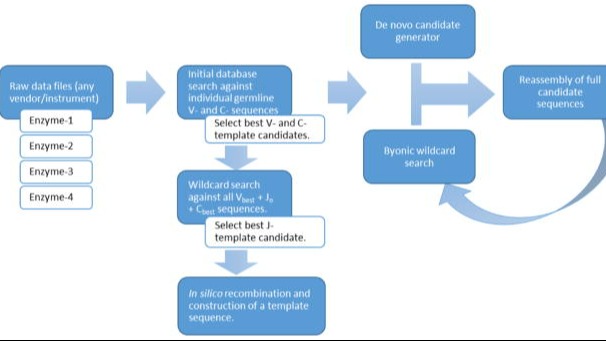
PUBLICATIONAutomated Antibody De Novo Sequencing and Its Utility in Biopharma Discovery
A novel method to automatically de novo sequence antibodies using mass spec and Byos, and a demonstration of algorithm robustness.
Byonic

VIDEO SERIESUsing Byonic
The Byonic tutorials explain MS/MS integration, peptide searches, charge adjustments, filtering PSMs, and report generation.

WEBINARSARS-CoV-2 Spike Ectodomain
Dive into HDX MS analysis of SARS-CoV-2 spike protein, focusing on glycopeptide inclusion and its challenges.

WEBINARBovine Milk Immunoglobulin G
Investigating the lactational differences in the N-glycosylation of bovine IgG and an overview of tools and methods including Byonic.

PUBLICATIONByonic: Advanced Peptide and Protein Identification Software
Byonic is a new software package for peptide and protein identification by tandem mass spectrometry, facilitating a wide range of search possibilities

PUBLICATIONPeak Filtering, Peak Annotation, and Wildcard Search for Glycoproteomics
Improvements to Byonic, a glycoproteomics search program, that addresses the analytical and bioinformatics challenges glycopeptide analysis poses.
Byosphere
VIDEO SERIESUsing Byosphere
Byosphere enhances LC-MS analysis with custom searches, dashboards, automated workflows, audit trails, and project management.

WEBINAREnhance Polyclonal IgG Product Characterization
CSL explore uses of Byosphere in characterizing polyclonal IgG products focusing on plasma product development.

WEBINARFrom Here To Byosphere
Heather Oakes discusses Lonza's CDMO mass spectrometry approach, focusing on flexibility for diverse molecules.

WEBINARDashboards By Deep Query
Use dashboards and deep query tools to analyze data, spot outliers, and enhance lab collaboration globally.
Japanese Resources

WEBINAR医薬品オリゴヌクレオチド分析の課題に対するソリューション
本セミナーでは、アジレントテクノロジーとプロテインメトリックスにおける、核酸医薬品LC,LCMS分析の最新ソリューションについてお話しします

WEBINAR効率的なMAM解析
タンパク質マトリックス MAM 分析プラットフォーム「Byos」および「Byosphere」をご覧ください。ライブデモやQ&Aも予定されています。

WEBINAR抗体関連医薬品の特性解析
LC-MS, icIEF-UV/MSを用いたチャージバリアント、糖鎖を含む詳細な修飾解析
Video Tutorials

VIDEO SERIESWorking with Intact Mass Workflows
Run intact mass workflows in Byos. Manage data, sequences, masses, deconvolution, and generate reports for ADCs, glycosylation, and oligos.

VIDEO SERIESWorking with Peptide Workflows
Explore peptide workflows, set up searches, interpret scores, validate peptides, manage disulfide mapping, and create reports.

VIDEO SERIESWorking with Chromatogram Workflows
Learn to use chromatogram workflows for peak analysis, adjust baselines, integrate peaks, align traces, and explore glycan analysis.

VIDEO SERIESWorking with Released Glycan Workflows
Learn to use released glycan workflows for N-glycans, O-glycans, and IGG glycans. Explore Glycan cartoons for visual representation.

VIDEO SERIESWorking with Oligonucleotide Workflows
Learn how to automate oligonucleotide data processing, handle raw files, define sequences, and generate custom reports.

VIDEO SERIESWorking with Digested Oligo Workflows
Learn Byos Digested Oligonucleotide workflows, modification setup, investigation tools, and custom report creation.

VIDEO SERIESWorking with Reconstruction Workflows
Perform intact reconstruction and charge variant analysis. Set up projects, configure processing nodes, and align CIEF traces effectively.

VIDEO SERIESWorking with Disulfides
Learn how to manage disulfide workflows, add samples, define sequences, create disulfide bridge and validate linked peptides.

VIDEO SERIESWorking with Hydrogen Deuterium Exchange (HDX)
The HDX tutorial series explains how to set up HDX workflows, analyze deuterium uptake, and generate detailed protein reports.

VIDEO SERIESWorking with Multi-Protein Quantitation
Rapidly analyze MS data to quantify proteins, manage workflows, and validate peptides in complex samples.

VIDEO SERIESUsing Byosphere
Byosphere enhances LC-MS analysis with custom searches, dashboards, automated workflows, audit trails, and project management.

VIDEO SERIESUsing Byonic
The Byonic tutorials explain MS/MS integration, peptide searches, charge adjustments, filtering PSMs, and report generation.














